Chapter 8 Interpretation
Load data
8.1 Variable importance
8.1.1 Random forest
# Created in 5-modeling.Rmd
base::load("data/model-rf.RData")
rf_imp = ranger::importance(rf)
# Sort by descending importance.
rf_imp = rf_imp[order(rf_imp, decreasing = TRUE)]
# Review top 10 real quick.
round(rf_imp, 4)[1:10]## thal_2 thal_3 oldpeak exang thalach ca_1 cp_2 sex_1 slope_2 ca_2
## 0.0344 0.0273 0.0271 0.0162 0.0157 0.0135 0.0131 0.0130 0.0108 0.0079## rf_imp[, drop = FALSE]
## thal_2 3.442978e-02
## thal_3 2.731880e-02
## oldpeak 2.707896e-02
## exang 1.620580e-02
## thalach 1.574323e-02
## ca_1 1.345678e-02
## cp_2 1.306690e-02
## sex_1 1.296439e-02
## slope_2 1.077906e-02
## ca_2 7.938321e-03
## slope_1 7.608534e-03
## age 7.582580e-03
## cp_1 5.827836e-03
## ca_3 4.018786e-03
## trestbps 3.535718e-03
## cp_3 3.075328e-03
## miss_cp 4.801077e-04
## restecg 3.609495e-04
## chol 3.543085e-04
## thal_1 6.954854e-05
## miss_thalach 5.743442e-05
## miss_exang 0.000000e+00
## fbs -2.573076e-06
## miss_fbs -1.383069e-05
## miss_sex -2.181920e-05
## miss_oldpeak -4.607897e-05
## ca_4 -9.516181e-05
## miss_ca -1.153608e-04
## miss_restecg -1.304717e-04
## miss_age -1.422576e-04
## miss_slope -1.479793e-04
## miss_chol -1.586197e-04
## miss_thal -2.750506e-04# Mean decrease in accuracy.
colnames(print_imp) = c("mean_dec_acc")
print_imp$var = rownames(print_imp)
print_imp2 = print_imp
# Add ranking to the rownames.
print_imp2$var = paste0(1:nrow(print_imp2), ". ", print_imp2$var)
print_imp2## mean_dec_acc var
## thal_2 3.442978e-02 1. thal_2
## thal_3 2.731880e-02 2. thal_3
## oldpeak 2.707896e-02 3. oldpeak
## exang 1.620580e-02 4. exang
## thalach 1.574323e-02 5. thalach
## ca_1 1.345678e-02 6. ca_1
## cp_2 1.306690e-02 7. cp_2
## sex_1 1.296439e-02 8. sex_1
## slope_2 1.077906e-02 9. slope_2
## ca_2 7.938321e-03 10. ca_2
## slope_1 7.608534e-03 11. slope_1
## age 7.582580e-03 12. age
## cp_1 5.827836e-03 13. cp_1
## ca_3 4.018786e-03 14. ca_3
## trestbps 3.535718e-03 15. trestbps
## cp_3 3.075328e-03 16. cp_3
## miss_cp 4.801077e-04 17. miss_cp
## restecg 3.609495e-04 18. restecg
## chol 3.543085e-04 19. chol
## thal_1 6.954854e-05 20. thal_1
## miss_thalach 5.743442e-05 21. miss_thalach
## miss_exang 0.000000e+00 22. miss_exang
## fbs -2.573076e-06 23. fbs
## miss_fbs -1.383069e-05 24. miss_fbs
## miss_sex -2.181920e-05 25. miss_sex
## miss_oldpeak -4.607897e-05 26. miss_oldpeak
## ca_4 -9.516181e-05 27. ca_4
## miss_ca -1.153608e-04 28. miss_ca
## miss_restecg -1.304717e-04 29. miss_restecg
## miss_age -1.422576e-04 30. miss_age
## miss_slope -1.479793e-04 31. miss_slope
## miss_chol -1.586197e-04 32. miss_chol
## miss_thal -2.750506e-04 33. miss_thalcolnames(print_imp2)[2] = "Variable"
# Reverse ordering of columns.
print_imp2 = print_imp2[, c(2, 1)]
rownames(print_imp2) = NULL
#
#print_imp2 = print_imp2[, c("Variable", "Mean Decrease Accuracy (%)")]#, "Description")]
#print_imp2 = print_imp2[, c("Variable", "\thead{Mean\\{}Decrease\\{}Accuracy (%)}")]#, "Description")]
print_imp2## Variable mean_dec_acc
## 1 1. thal_2 3.442978e-02
## 2 2. thal_3 2.731880e-02
## 3 3. oldpeak 2.707896e-02
## 4 4. exang 1.620580e-02
## 5 5. thalach 1.574323e-02
## 6 6. ca_1 1.345678e-02
## 7 7. cp_2 1.306690e-02
## 8 8. sex_1 1.296439e-02
## 9 9. slope_2 1.077906e-02
## 10 10. ca_2 7.938321e-03
## 11 11. slope_1 7.608534e-03
## 12 12. age 7.582580e-03
## 13 13. cp_1 5.827836e-03
## 14 14. ca_3 4.018786e-03
## 15 15. trestbps 3.535718e-03
## 16 16. cp_3 3.075328e-03
## 17 17. miss_cp 4.801077e-04
## 18 18. restecg 3.609495e-04
## 19 19. chol 3.543085e-04
## 20 20. thal_1 6.954854e-05
## 21 21. miss_thalach 5.743442e-05
## 22 22. miss_exang 0.000000e+00
## 23 23. fbs -2.573076e-06
## 24 24. miss_fbs -1.383069e-05
## 25 25. miss_sex -2.181920e-05
## 26 26. miss_oldpeak -4.607897e-05
## 27 27. ca_4 -9.516181e-05
## 28 28. miss_ca -1.153608e-04
## 29 29. miss_restecg -1.304717e-04
## 30 30. miss_age -1.422576e-04
## 31 31. miss_slope -1.479793e-04
## 32 32. miss_chol -1.586197e-04
## 33 33. miss_thal -2.750506e-04# Convert to a percentage.
print_imp2[, 2] = print_imp2[, 2] * 100
# TODO: fix variable names and restrict to ~15 variables.
# Manually escape variable names.
print_imp2$Variable = gsub("_", "\\_", print_imp2$Variable, fixed = TRUE)
print_imp2## Variable mean_dec_acc
## 1 1. thal\\_2 3.4429779088
## 2 2. thal\\_3 2.7318796325
## 3 3. oldpeak 2.7078957923
## 4 4. exang 1.6205799641
## 5 5. thalach 1.5743230136
## 6 6. ca\\_1 1.3456775599
## 7 7. cp\\_2 1.3066898914
## 8 8. sex\\_1 1.2964385593
## 9 9. slope\\_2 1.0779059934
## 10 10. ca\\_2 0.7938320939
## 11 11. slope\\_1 0.7608534071
## 12 12. age 0.7582579788
## 13 13. cp\\_1 0.5827836422
## 14 14. ca\\_3 0.4018786023
## 15 15. trestbps 0.3535717660
## 16 16. cp\\_3 0.3075327860
## 17 17. miss\\_cp 0.0480107702
## 18 18. restecg 0.0360949501
## 19 19. chol 0.0354308487
## 20 20. thal\\_1 0.0069548539
## 21 21. miss\\_thalach 0.0057434419
## 22 22. miss\\_exang 0.0000000000
## 23 23. fbs -0.0002573076
## 24 24. miss\\_fbs -0.0013830689
## 25 25. miss\\_sex -0.0021819198
## 26 26. miss\\_oldpeak -0.0046078973
## 27 27. ca\\_4 -0.0095161808
## 28 28. miss\\_ca -0.0115360833
## 29 29. miss\\_restecg -0.0130471721
## 30 30. miss\\_age -0.0142257555
## 31 31. miss\\_slope -0.0147979304
## 32 32. miss\\_chol -0.0158619663
## 33 33. miss\\_thal -0.02750506478.2 Accumulated local effect plots
task = makeClassifTask(data = data[, c(vars$predictors, vars$outcomes[1])],
target = vars$outcomes[1])
learner = makeLearner("classif.ranger",
predict.type = "prob",
# TODO: confirm best mtry.
mtry = 4,
num.trees = 200L,
num.threads = get_cores())
# This takes 1 second
system.time({
mod.mlr = mlr::train(learner, task)
})## user system elapsed
## 0.03 0.01 0.03## Ranger result
##
## Call:
## ranger::ranger(formula = NULL, dependent.variable.name = tn, data = getTaskData(.task, .subset), probability = (.learner$predict.type == "prob"), case.weights = .weights, ...)
##
## Type: Probability estimation
## Number of trees: 200
## Sample size: 303
## Number of independent variables: 33
## Mtry: 4
## Target node size: 10
## Variable importance mode: none
## Splitrule: gini
## OOB prediction error (Brier s.): 0.1339879mod2 = Predictor$new(mod.mlr, data = data[, !names(data) %in% vars$outcomes[1]],
y = data[[vars$outcomes[1]]])
# Default plot
#effect$plot()
# Calculate multiple ALE effects at a time.
system.time({
effect2 = iml::FeatureEffects$new(mod2,
grid.size = 20L,
features = c("oldpeak", "trestbps"))
})## Warning: package 'ellipsis' was built under R version 3.6.3## Warning: package 'lubridate' was built under R version 3.6.3## Warning: package 'xml2' was built under R version 3.6.3## Warning: package 'pROC' was built under R version 3.6.3## Warning: package 'Metrics' was built under R version 3.6.3## Warning: package 'backports' was built under R version 3.6.3## Warning: package 'glue' was built under R version 3.6.3## Warning: package 'reshape2' was built under R version 3.6.3## Warning: package 'parallelMap' was built under R version 3.6.3## Warning: package 'vctrs' was built under R version 3.6.3## Warning: package 'openxlsx' was built under R version 3.6.3## Warning: package 'lifecycle' was built under R version 3.6.3## Warning: package 'foreach' was built under R version 3.6.3## Warning: package 'lava' was built under R version 3.6.3## Warning: package 'rlang' was built under R version 3.6.3## Warning: package 'ROCR' was built under R version 3.6.3## Warning: package 'purrr' was built under R version 3.6.3## Warning: package 'prediction' was built under R version 3.6.3## Warning: package 'recipes' was built under R version 3.6.3## Warning: package 'tidyselect' was built under R version 3.6.3## Warning: package 'plyr' was built under R version 3.6.3## Warning: package 'bookdown' was built under R version 3.6.3## Warning: package 'pillar' was built under R version 3.6.3## Warning: package 'haven' was built under R version 3.6.3## Warning: package 'withr' was built under R version 3.6.3## Warning: package 'RCurl' was built under R version 3.6.3## Warning: package 'tibble' was built under R version 3.6.3## Warning: package 'future.apply' was built under R version 3.6.3## Warning: package 'forcats' was built under R version 3.6.3## Warning: package 'ModelMetrics' was built under R version 3.6.3## Warning: package 'tidyr' was built under R version 3.6.3## user system elapsed
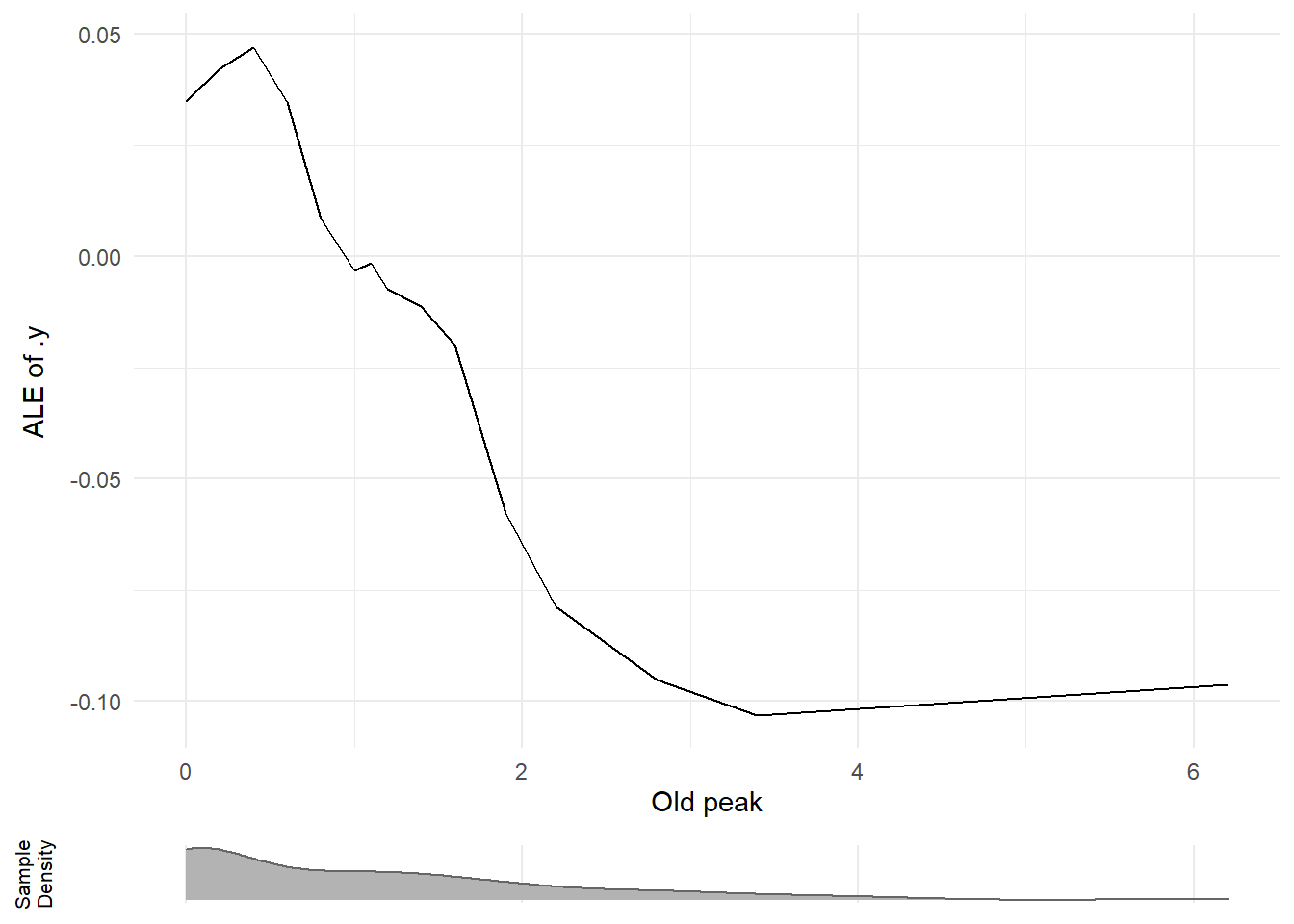
## 1.30 0.21 1.45
# Improved plot
# TODO: move into ck37r
plot_ale = function(var, var_display) {
if (FALSE) {
effect = iml::FeatureEffect$new(mod2,
grid.size = 32L,
feature = var)
old_results = effect$results
effect$results = old_results[old_results$.class == 1, ]
}
old_results = effect2$effects[[var]]$results
effect2$effects[[var]]$results = old_results[old_results$.class == 1, ]
(p = effect2$effects[[var]]$plot(rug = FALSE) +
theme_minimal() + theme(strip.text.x = element_blank()) +
labs(x = var_display))
# Density plot
(p2 = ggplot(data = data, aes_string(x = var)) +
geom_density(fill = "gray70", color = "gray40") +
theme_minimal() + labs(y = "Sample\nDensity") +
#scale_x_log10(breaks = breaks, limits = limits) +
theme(axis.title.x = element_blank(),
axis.title.y = element_text(size = 8),
# Include x-axis major gridlines to ensure that plots are aligned.
panel.grid.major.y = element_blank(),
panel.grid.minor = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_blank()))
print(cowplot::plot_grid(p, p2, align = "v", ncol = 1, rel_heights = c(0.9, 0.1)))
ggsave(paste0("visuals/ale-", var, ".pdf"), width = 4, height = 4)
}
plot_ale("oldpeak", "Old peak")